Research focus
The Wu Lab focuses on the integration and analysis of omics data (genome, transcriptome, proteome, metabolome, macrogenome, etc.), especially the research of metagenomics. The Wu Lab uses machine learning, deep learning, artificial intelligence and causal inference algorithms for development and application. And we are also engaged in computer-aided drug design, including TCM informatics and TCM modernization research.
Targeting keystone species helps restore the dysbiosis of butyrate-producing bacteria in nonalcoholic fatty liver disease
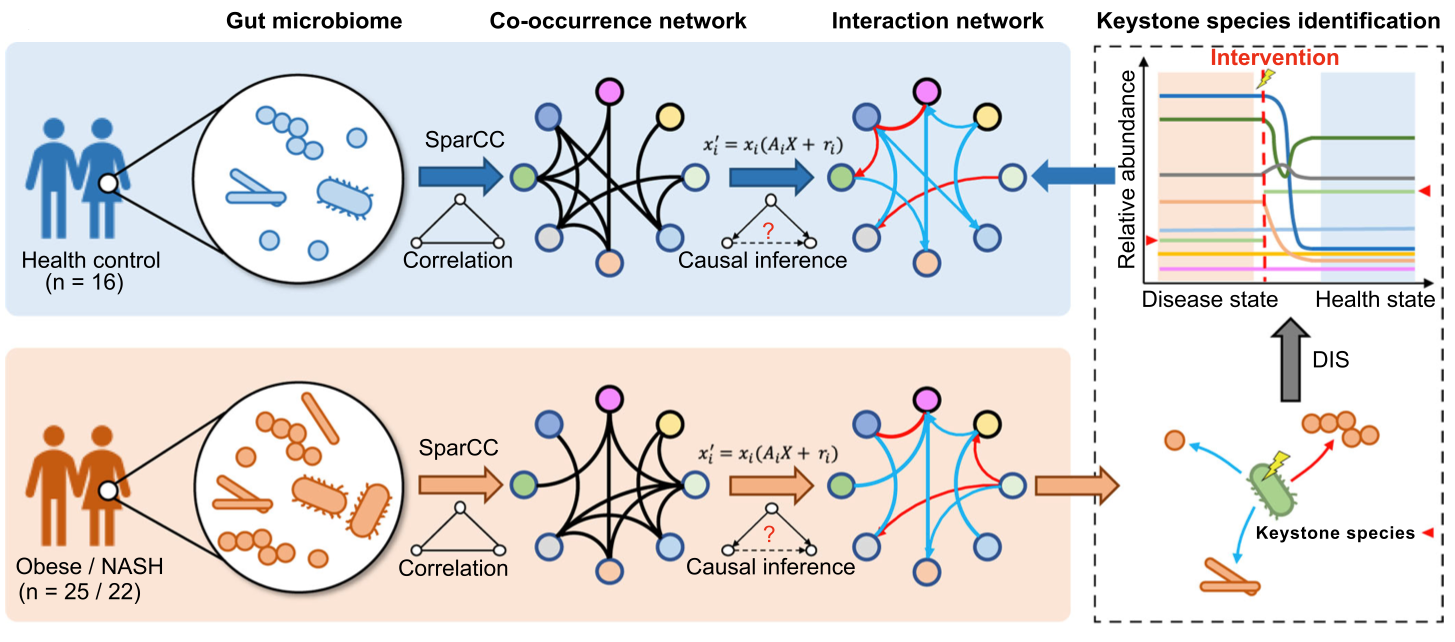
The dysbiosis of the gut microbiome is one of the pathogenic factors of nonalcoholic fatty liver disease (NAFLD) and also affects the treatment and intervention of NAFLD. Among gut microbiomes, keystone species that regulate the integrity and stability of an ecological community have become the potential intervention targets for NAFLD. An algorithm was implemented to identify keystone species based on causal inference theories and dynamic intervention simulation. Our findings demonstrate the importance of keystone species in restoring the microbial composition toward a normal gut microbiome, suggesting a novel potential microbial treatment for NAFLD.
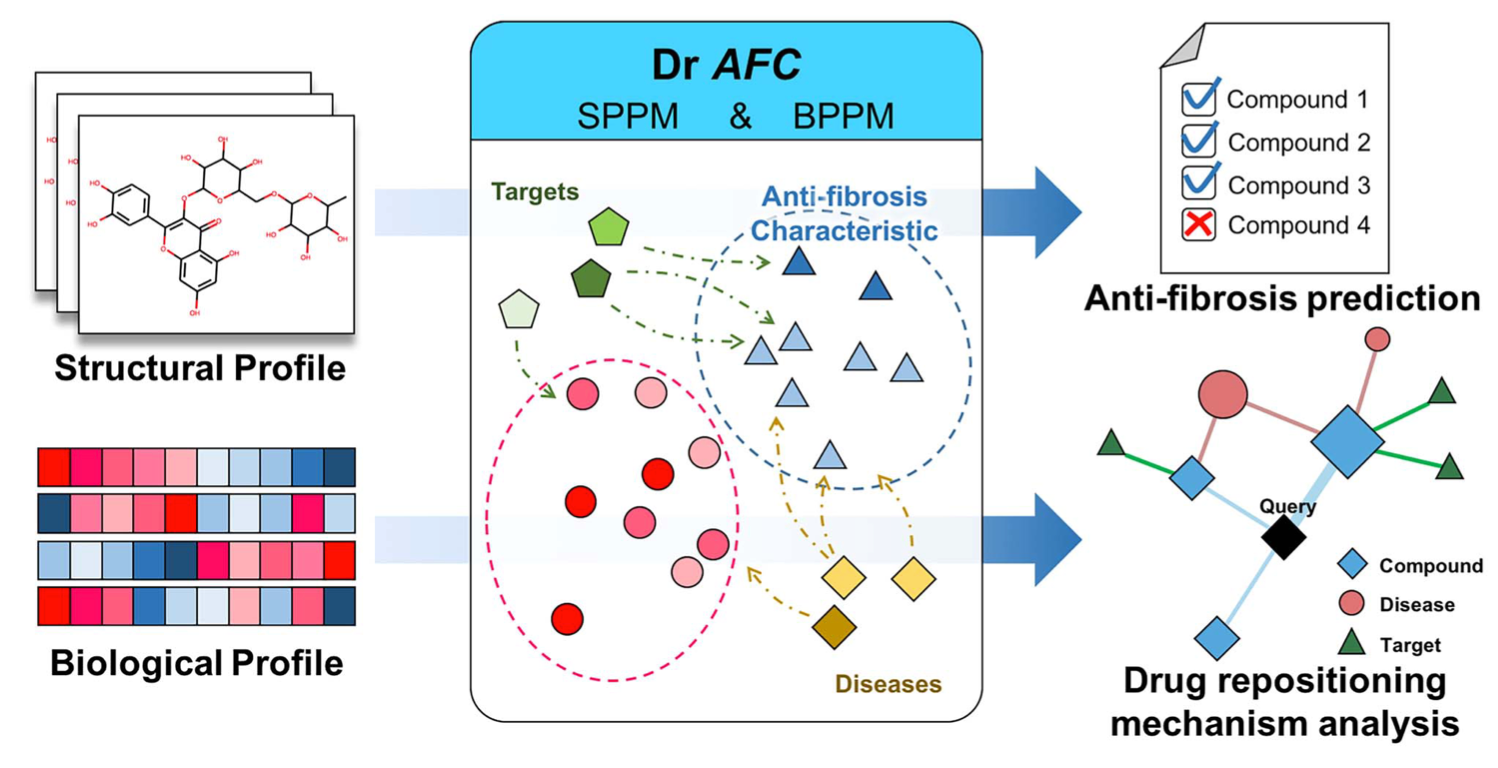
Dr AFC: drug repositioning through anti-fibrosis characteristic
In this study, anti-fibrosis knowledge base was first built by collecting data from multiple resources. Both structural and biological profiles were then derived from the knowledge base and used for constructing machine learning models including Structural Profile Prediction Model (SPPM) and Biological Profile Prediction Model (BPPM). Three external public data sets were employed for validation purpose and further exploration of potential repositioning drugs in wider chemical space. The resulting SPPM and BPPM models achieve area under the receiver operating characteristic curve (area under the curve) of 0.879 and 0.972 in the training set, and 0.814 and 0.874 in the testing set. Web server: https://www.biosino.org/drafc.
Phylogenetic analyses with systematic taxon sampling show that mitochondria branch within Alphaproteobacteria
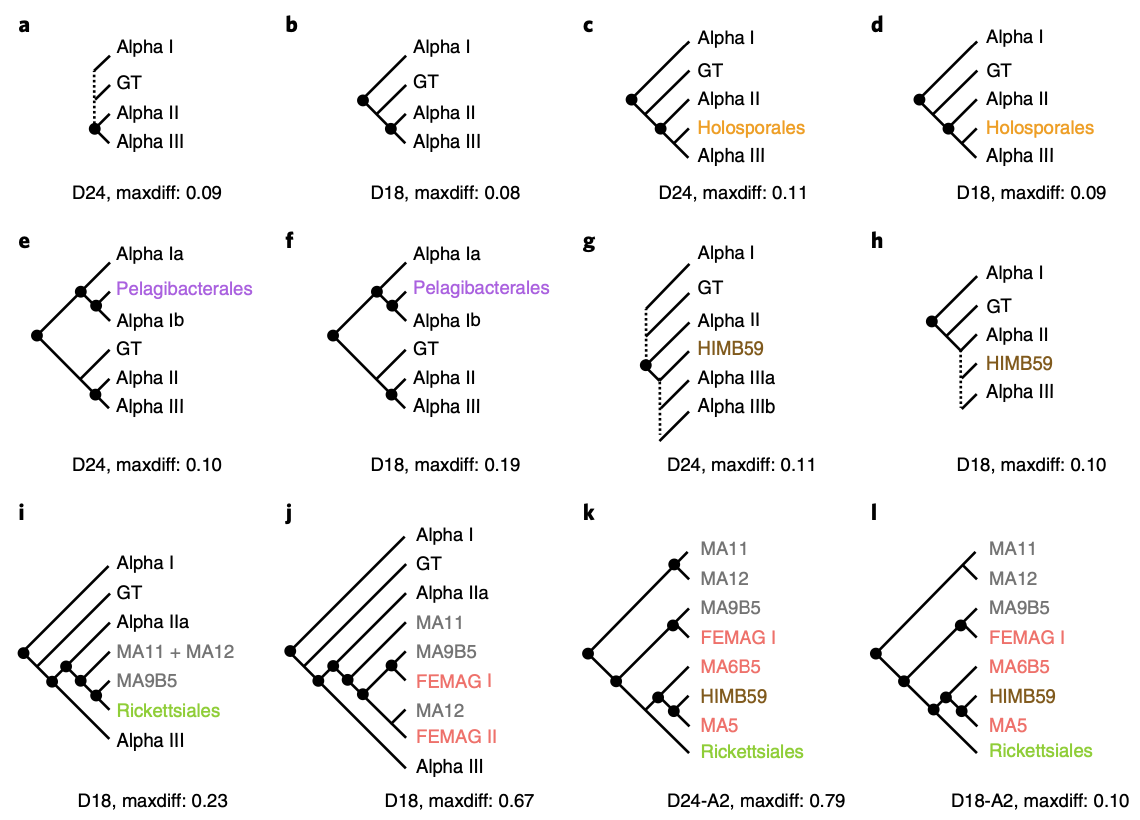
Though it is well accepted that mitochondria originated from an alphaproteobacteria-like ancestor, the phylogenetic relation- ship of the mitochondrial endosymbiont to extant Alphaproteobacteria is yet unresolved. The focus of much debate is whether the affinity between mitochondria and fast-evolving alphaproteobacterial lineages reflects true homology or artefacts. We demonstrate that site-exclusion methods produce erratic phylogenetic estimates of mitochondrial origin. Thus, previous phylogenetic hypotheses on the origin of mitochondria based on pretreated datasets should be re-evaluated. We applied alternative strategies to reduce phylogenetic noise by systematic taxon sampling while keeping site substitution information intact. Cross-validation based on a series of trees placed mitochondria robustly within Alphaproteobacteria, sharing an ancient common ancestor with Rickettsiales and currently unclassified marine lineages.